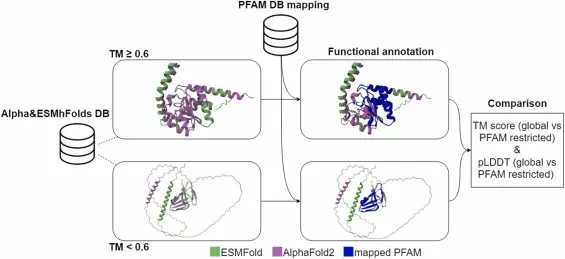
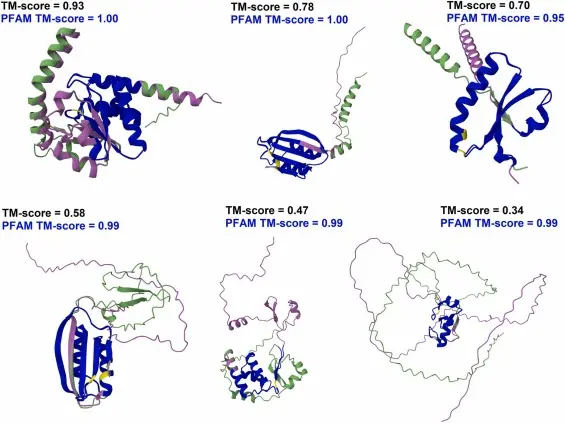
In recent years, artificial intelligence (AI) has brought a revolution to the world of biochemistry — particularly in how we understand the 3D structure of enzymes, the biological catalysts that power nearly every chemical reaction in the human body.
Understanding enzyme structures is critical. Their shape determines their function, and changes to that shape can affect health, disease, drug response, and metabolic pathways. Traditionally, revealing these structures required slow and expensive experimental techniques, such as X-ray crystallography or cryo-electron microscopy. But now, with AI, we can predict these structures in minutes, with high accuracy.
🔍 Why Predicting Enzyme Structures Matters
Enzymes are proteins that speed up chemical reactions. In humans, they play essential roles in:
- Digestion and metabolism
- DNA repair
- Immune response
- Signal transduction
- Detoxification and drug metabolism
When enzymes malfunction due to mutations, misfolding, or inhibition it can lead to serious health conditions, from metabolic disorders to cancer. That's why being able to predict and visualize enzyme structures is vital for medicine, biotechnology, and basic research.
🧠 Enter AlphaFold2 and ESMFold
Two AI tools have made headlines for their ability to predict protein structures:
- AlphaFold2 (DeepMind): An advanced AI that uses evolutionary data to predict 3D protein structures with atomic-level accuracy. It’s been described as one of the biggest scientific breakthroughs of the decade.
- ESMFold (Meta AI): A newer method that uses a protein language model instead of evolutionary comparisons. It’s significantly faster and does not require large multiple sequence alignments.
Both tools are open-access and freely available to researchers — making cutting-edge structure prediction accessible worldwide.
⚙️ From Sequences to Structure — in Minutes
These AI systems take an amino acid sequence — essentially, the "letters" that make up a protein — and use deep learning to predict how the chain will fold into a complex 3D shape. This folded structure determines how the enzyme interacts with other molecules, substrates, or drugs.
Thanks to this technology, researchers can now:
- Predict structures for entire enzyme families
- Explore unknown or uncharacterized proteins
- Design mutations for enzyme engineering
🧪 Real-World Applications
- Medicine: Predicting how disease mutations change enzyme shape and function
- Pharmaceuticals: Designing drugs that bind better to enzyme targets
- Synthetic Biology: Engineering enzymes to work in new environments
- Diagnostics: Developing biosensors and enzyme-based assays
We are entering a new era where AI doesn’t just assist research — it drives it. For enzyme biology, this means faster discoveries, more personalized medicine, and innovative biotech applications.
Whether you're studying enzymes in health, designing novel biocatalysts, or mapping molecular pathways — tools like AlphaFold2 and ESMFold are changing what’s possible.